Back to BioTapestry Home Page
Release Notes for Version 2.1
Version 2.1 was released on 7/23/07. It includes two new significant features: optional 2D model images, and user-specified
paths through the model hierarchy. These features are highlighted in this screen shot:
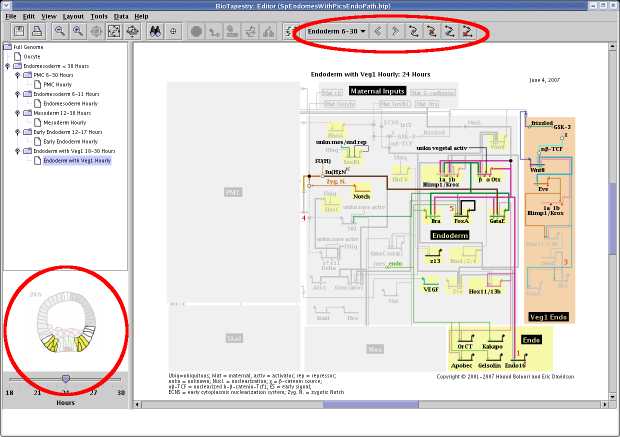
-
Display of a 2D view with each network model: A PNG, GIF, or JPEG image can be assigned
to each different model in the hierarchy, including each separate time point in
a dynamic model; this image is then displayed in the lower left of the application
when the model is viewed. Note: Since this feature can quickly increase
the size of your saved model file, it is recommended to display thumbnail-sized
pictures (e.g. 150 x 150 pixels). To access this feature, select:
Edit->Assign Image to Current Model...
-
User paths: You can now specify a series of models in the hierarchy
as a "user path", with each model as a stop on the path. This allows you to
quickly and smoothy navigate through those models, even if they are scattered throughout
the model hierarchy. This can be used, for example, to track the development of
a particular cell lineage through time. Note: you need to have at least two models defined
before the feature is activated. To access this feature, click on
the Create New Path button on the toolbar, or select:
View->Manage Navigation Paths->Create New Path
Then, navigate to each successive model you want to include in the path and click on
the Add New Stop to Path button on the toolbar, or select:
View->Manage Navigation Paths->Add New Stop to Path
Once you have created one or more paths, you can select each path using the
path choice selector in the toolbar, and then using the Step Forward Along Path
or Step Back Along Path toolbar buttons. Or you can select the path from the
View->Manage Navigation Paths->Select Current Path submenu,
and then select:
View->Manage Navigation Paths->Step Forward Along Path
to move forward. A similar menu item allows you to step backward.
Release Notes for Version 2.0.2
Version 2.0.2 was released on 6/14/07. Changes include:
- More robust error checking and diagnostics for file input and output
- User is asked before overwriting existing files on most output operations
- Added an "Open Recent Files" menu
- Added finer-grained zoom settings to optimize sizing with "Zoom to Model" action
- Bug fix for rendering intercellular nodes with variable activity levels
- Code is signed with new security certificates
Release Notes for Version 2.0.1
Version 2.0.1 was released on 5/22/07. Changes include:
- Fixed problem reading and writing files on Windows with Java 1.6
- Changed URL to www.BioTapestry.org in the HelloWebPlugIn example
- Notes associated with expression profiles now appear in the experimental data window
Release Notes for Version 2.0
Version 2.0 was released on 5/17/07. It includes a number of significant new features:
-
Improved Automatic Layout: The user can select between two types of automatic layout
"templates": General or Bipartite.
-
The General layout can handle any type of topology. All genes with outputs
are grouped on the left, while pure targets are grouped on the right. The criteria
for ordering the groups can be set as layout options (e.g. time of first expression,
topological sort of the network, common inputs, etc). Ancillary nodes such as bubbles
or signals are clustered with the most appropriate source or target gene. To access
this feature, select:
Layout->Apply Auto Layouts->General Strategy
-
The Bipartite layout works well when genes can be
divided into two disjoint groups: sources and targets. In other words, source genes
cannot themselves be targets of other source genes. While obviously very specialized,
it is very useful for a certain class of data sets. It can handle networks that
are not strictly bipartite, in that certain ancillary nodes (bubbles, signals, etc.) that
are themselves targets, but which then have outputs which are exclusive to a target gene,
will be laid out adjacent to the target gene. To access this feature, select:
Layout->Apply Auto Layouts->Bipartite Strategy
-
These new layouts are used when importing SIF files or CSV files, unless the
user requests that the current layout be maintained. Layout parameters for
these imports can be set using the layout options dialog:
Layout->Set Automatic Layout Options
-
Improved Link Rendering: There is much more flexibility in customizing
how links get drawn. Link rendering properties, including color, thickness,
and style, can be specified either for individual link segments, for individual
links in a tree of links, or for the entire tree of links. In the case of
per-link properties, those properties will be retained and applied as needed
even when link trees are laid out differently. To access the improved link rendering
feature, right-click on a link and select Properties..., and select the
Presentation Properties tab. Per-link properties can be set by selecting
a listed link in the table and clicking on Edit Entry....
-
Configurable Workspace Size: The size of the workspace is no longer
fixed; the user can specify sizes up to very large values, allowing
rather large networks to be modeled. To access this feature, select:
Layout->Set Workspace Size...
-
User-Configurable Time Units: User can specify the time units to use,
ranging from seconds to years, or can specify numbered or custom named stages.
To access this feature, select:
Data->Set Time Units...
-
New Node and Link Activity Level Features:
-
The activity level of nodes and links in submodels are no longer just on or off:
they are fully variable. The activity level of nodes can be depicted
using, color saturation, a small pie glyph, or both.
The activity level of links, on a per-link basis within a tree of links,
can be depicted using color saturation, line thickness, or both. Link levels
can be tied to the source node, or set separately. (Note that this feature
is not available at the root model Full Genome level.)
To access the variable activity feature, right-click on a node in a submodel,
select Properties..., set the Activity option to Variable,
and fill in a value between 0.0 and 1.0. For links, also right-click on the link
in a submodel, select Properties..., and set the Activity option for
the desired link in the table to either Specify or Source level. With
the former setting, you need to fill in a value between 0.0 and 1.0. With the
latter setting, the link will match the activity level of the source node.
To select the display method used for showing the variable activity, select:
Edit->Set Display Options...
and then set the Node Activity Display and Link Activity Display options.
-
The variable activity level feature is now being used in dynamic submodels to depict
the "weak" expression level setting in the time course data (fully variable expression
levels in the time course data have not yet been implemented). The activity
level assigned to the "weak" expression level is set by selecting:
Edit->Set Display Options...
and then entering a value between 0.0 and 1.0 in the Expression level for weak
expression field.
-
Improved Zoom Features: New zoom options have been introduced, including
zoom to selection, zoom to show all of current model, zoom to bound all
models in the hierarchy, and zoom to show full workspace. Most of these options
are available as buttons on the toolbar; all options are available as selections
in the main View menu.
-
Improved Navigation Options: The user can now specify which model is displayed
at startup, what zoom level to use at startup, and how to change zoom levels when
switching between model views. The model to display at startup is chosen by
right-clicking on a model in the left-hand navigation panel, then selecting
Make This Model the View on Startup. The other options are set by
selecting:
Edit->Set Display Options...
and then setting the Set Initial Zoom on Load and Zoom When Selecting options.
-
Insert Nodes into Preexisting Links: The user can now
insert nodes or genes into an existing linkage in the root model,
and it will be automatically propagated to all submodels that contain that link.
To do this, right-click on a link in the root model only,
select Insert Node into Link or Insert Gene into Link.
-
User-Specified Link Pad Counts for Most Node Types: Previous versions
only allowed genes to have extra link landing pads. Now, all
node types but slashes can have extra link pads added. To do this, right-click
on a node, choose Properties..., check the Add extra length box,
and select the pad count you desire.
-
Arbitrary Text as Experimental Data: You can now display
arbitrary simple HTML text when you right-click on a gene
and select Experimental Data. To enter this text, right-click
on a gene, choose Properties..., and select the Text Annotations tab.
You can enter simple formatting tags, e.g., italics, bold, paragraphs, and line breaks.
-
Improved Model Query and Search:
-
The search dialog has been improved, with new options such as
partial name matching, simultaneous link selection,
and zooming to selections following the search. Click on the search
button on the toolbar, or select Tools->Search....
-
When right-clicking on a link segment, you can choose to select and zoom to
the link source node, or alternately to select all targets through the
segment and zoom to them. From the right-click menu, either choose
Select Link Source or Select Link Targets Through This Segment.
-
"Build Network From Description" Dialog Improvements: The dialogs for
building a network automatically from a list of interactions have some improvements.
-
The tabs and tables have been rearranged so that all standard interactions now
appear in one table under the Standard Interactions tab, where
the user can select the appropriate level of table complexity for what they need.
-
Standalone nodes are now supported (i.e. nodes that have no associated links).
These are specified in using the Standalone Nodes tab.
-
If the type of a node (e.g. gene, bubble) has already been specified for a node,
subsequent uses of that name will automatically set the type selection.
-
When the network is laid out the first time, or if the layout is discarded, the
new template layout types will now be used. The old layout algorithm is only
used for incremental additions to the network.
-
Standalone Nodes are Supported in CSV Inputs: The CSV import now supports
a "nodeOnly" keyword, e.g:
"# Command Type","Model Name","Node Type","Node Name","Region Abbrev"
"nodeOnly","Tutorial Model","gene","Gene 2","A"
-
New Link Confidence Display Options: The ways to graphically display the
experimental confidence for a link have been enhanced. In addition to three
different diamond colors at the link target, the user can choose to use the
link thickness to display the confidence. To set the display method, select:
Edit->Set Display Options...
and choose the desired Link Evidence Display option. The evidence for
a link is set using by right-clicking on the link and setting the Cis-Reg Evidence
level in the table. The three available levels have no predefined semantics.
-
New Network Duplication Features: Features have been added that allow the
user to create copies of parts of existing networks.
-
A submodel, and all its child models, can be duplicated as a new sibling
submodel in the model hierarchy. Right-click on a submodel in the left-hand
navigation panel, and choose Duplicate Model as Sibling
-
All the nodes and links in a region of a submodel can be duplicated into
a new region of the submodel. In a submodel, right click on a region rectangle
and select Duplicate Region
-
New Automatic Layout Tools: Some new tools help to do layout:
-
The link and node colors can be reassigned to eliminate crossing ambiguities.
Select:
Layout->Other Automatic Layout Tools->Reassign Colors to Remove Ambiguity
-
Non-orthogonal links can be selected, allowing the user to track down links that may need tweaking.
Select:
Layout->Other Automatic Layout Tools->Select Non-Orthogonal Link Segments
-
There are tools to center both the current network model, as well as all the network models, in the
workspace. Also, all the different network models can be aligned witch each other. These tools are
found under:
Layout->Layout Centering and Alignment
-
Lots of Other Small Improvements:
-
The ordering of the models in the left-hand navigation display can be changed. Right-click on a model
name and select Swap Position with Previous Model or Swap Position with Next Model to
change the ordering among sibling models in the model hierarchy.
-
A Cancel button has been added to all long-running operations.
-
The model stays centered when the window is resized.
-
Color selection choices now display color swatches, and colors are ordered by hue.
-
Link drawing in the view-only application is faster.
-
Slash nodes can now be oriented in all directions. Right-click on a slash node,
select Properties..., select the Presentation Properties tab, and set
the Orientation.
-
The size of the repression link "feet" can be increased by the user. Select:
Edit->Set Display Options...
and type a positive integer into the Extra Repression Bar Length field.
-
Right-click on a link, select Swap Link Pads, and click on a link pad; the
current occupant of the new pad will swap positions with the current pad. (Note that
since a link source pad cannot be shared with a link target pad, this is the only
way to reassign the link pads on slash nodes, which have only two pads.)
-
A new comma-separated value (CSV) format for importing raw QPCR data has
been added to the original format.
-
You can choose Edit->Select None to unselect everything in the current selection.
-
In addition to displaying model date and attribution data, the user can define and
place a block of model key text. Select Data->Edit Model Description Data... and
type into the Key Text field.
-
The data structures used to represent unaffected genes in the QPCR data tables are now
much more robust. Futhermore, newly imported QPCR data causes the status of unaffected
genes to be automatically re-evaluated and recategorized if appropriate.
-
The name of a non-gene node can be hidden in the network display. Right-click on
the node, select Properties..., and select the Presentation Properties tab.
Then check the Hide node name box.
-
The current open file name is displayed in the window bar.
-
Submodels can now be tagged with time bounds (e.g. minimum and maximum hours).
-
Several bug fixes have been folded into the code.
Back to BioTapestry Home Page
Last updated: September 5, 2023
biotapestry at systemsbiology dot org






