Back to BioTapestry Home Page
The Gaggle was a framework, developed at the Baliga Lab of the Institute for Systems Biology, "...for exchanging data between independently
developed software tools and databases to enable interactive exploration of systems biology data." The BioTapestry Editor supported Gaggle through the 7.0.0 release, and
provided a Java WebStart link on this page to launch a Gaggle-enabled BioTapestry Editor. We no longer provide WebStart launches, and that offering has been retired. The
instructions below are retained for historical purposes.
After first launching the Gaggle Boss, click HERE (formerly a Java WebStart link, now retired) to run the Gaggle-enabled BioTapestry
Editor (version 7.0.0, released 9/22/14, requires Java 1.5 or above) using Java Web Start. Since the Gaggle Boss and the provided Gaggle libraries are self-signed,
using this tool will typically involve adding security execptions for the sites http://gaggle.systemsbiology.net/2007-04/boss.jnlp and
http://www.biotapestry.org/webStart/gaggle.jnlp via the Java Control Panel Security tab on your computer.
Using BioTapestry with Gaggle
Here is a list of essential points to know when using BioTapestry with Gaggle:
- The Gaggle Boss must be running before launching BioTapestry.
- You can launch the Boss and other Gaggle-aware applications using the links on the Gaggle Blank Slate (formerly a link to a now-retired page).
- You will need to accept a self-signed security certificate from Paul Shannon to be able to use the Gaggle libraries (unless you have
already specified to always trust that certificate).
- The Gaggle species that BioTapestry uses to tag data broadcasts is hardwired to
unknown on startup, though inbound
data broadcasts will change this.
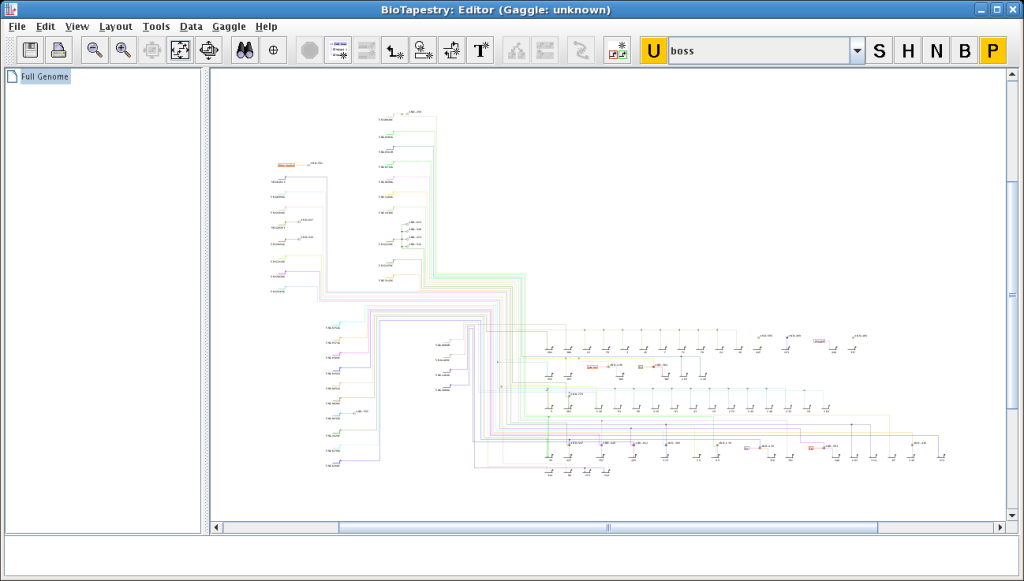
- Unlike other Gaggle-aware applications, incoming data are not immediately incorporated automatically into the network model. Instead, BioTapestry has a
special Process Pending Gaggle Requests button (labeled P) in the Gaggle control set that turns orange when new data is available; click on that button
to pull in the new data. In a similar fashion, there is an Update Goose List button (labeled U) which turns orange and must be clicked when
the list of available geese needs to be updated:
- BioTapestry accepts only two Gaggle data types: networks and lists. When sent a network, BioTapestry throws away the entire existing full model hierarchy (if any) and creates an
entirely new single Full Genome model. (There is no support for building a model hierarchy via Gaggle inputs.) When sent a list, BioTapestry treats it as a list of node
names to select in the currently displayed network. The selections are added to any current selections, and a name is ignored if the node does not exist in the model. All nodes with the given name are selected (e.g. If the current model is a submodel with multiple regions, each node copy with the name is selected).
- You can only transmit a network from the top-level Full Genome model. When sending out a network, BioTapestry sends out the currently selected
elements (nodes and links). If a link is selected, the source and target nodes are implicitly included.
- When transmitting a list (possible at any model level), the names of the currently selected nodes are sent in the list.
- If nodes and edges of an inbound network have been annotated, BioTapestry can assign node types and link signs. If no such annotation exists, every node will be created
as a gene, and every link will have a positive sign. Here is a tiny example of a correct node attributes
.noa file:
NodeType
VNG5182G = gene
VNG1405C = gene
AND-665 = bubble
gamma = box
139 = gene
Acceptable tags are: bare, box, bubble, intercel,
slash, diamond, and gene.
And here is a tiny example of a correct edge attributes .eda file:
LinkSign
VNG1405C (activates) 139 = positive
gamma (combines) AND-665 = positive
VNG5182G (combines) AND-665 = positive
AND-665 (represses) 139 = negative
Acceptable tags are: positive, negative, and neutral.
- Important notes about automatic layout strategy:
- When BioTapestry receives a network from Gaggle, it uses the current "initial" automatic layout algorithm policy to layout the network. To set this policy, select
Layout->Set Automatic Layout Options from the main menu, and then select the Initial Layouts tab in the dialog. The two supported
strategies are the Bipartite Strategy and the General Strategy. These two strategies are described in the BioTapestry FAQ
here. The default behavior is to try to apply the bipartite strategy
first. This strategy requires that every gene in the network must be either a link source or a link target; no gene can be both or neither. If this requirement
is not met, the system falls back to use the general strategy. Note that this means that a standalone node (no inputs) in the incoming network will force
the selection of the general strategy!
- One important feature of the "bipartite" strategy should be kept in mind; the network does not have to be formally bipartite
in that if there are non-gene (e.g. bubble) nodes that are associated exclusively with a single target gene, they are grouped with that target gene in the layout. Note that
this means you must specify node attributes for these nodes before they are broadcast over the Gaggle if you want them to be handled
correctly to trigger the use of the bipartite layout.
- Even if the system rejects the network for not meeing the requirements for a bipartite layout (e.g. it has standalone genes), you can force it by
selecting Layout->Apply Auto Layouts->Bipartite Strategy from the main menu. You just need to select Yes when the dialog pops
up and asks "This layout cannot handle all nodes in the network. Continue anyway?". This message just means that whatever genes or nodes
don't meet the topology requirements for the bipartite layout algorithm will be left untouched while the rest are handled. You will need to move the non-conforming
nodes and links around by hand. Note that the general layout algorithm always has a way to handle all nodes and links; only the bipartite strategy has
network topology restrictions.
Back to BioTapestry Home Page
Last updated: September 5, 2023
biotapestry at systemsbiology dot org






