|
|
| News |
|---|
| September 25, 2023: New Version 7.1.3 has been released just for the Mac. See What's New. |
| September 28, 2019: New Version 7.1.2 has been released. See What's New. |
BioTapestry is an interactive tool for building, visualizing, and sharing gene regulatory network models over the web.
Downloads
The latest version of the BioTapestry Editor for Windows and Linux is version 7.1.2, released 9/28/19. For Macs, the latest version is now version 7.1.3, released 9/25/2023.
Download the copy of BioTapestry that is appropriate for your system. In contrast to previous versions, you no longer need to download and install Java first; these downloads now include the required Java files, and so the program is standalone.
If you need to use an older version of BioTapestry prior to Version 7.1.2 and 7.1.3, you can still download older versions of the program
from the Archived Executables page. Depending on the version and operating system, you may need
to install Java as well.
- Windows: The Windows version now comes with an installer program that also installs the Java runtime.
Zip archive: BioTapestry7120.zip Download the archive and unzip it as a folder on
your desktop. Go into the folder, and double-click the
BioTapestryInstaller program. If you want to install BioTapestry
in the Windows "Program Files" folder, you will need to run the BioTapestryInstaller as an Admin user. To do that, right-click on
the BioTapestryInstaller program and choose "Run as administrator". You will be prompted for the Administrator password.
If you are not an Admin, you will need to install it somewhere within your home directory by changing the install location as part of the
installation. Once the installation is complete, you can right-click on the BioTapestry program and select "Create shortcut", then
e.g. drag it to your desktop. MD5 checksum: MD5 (BioTapestry7120.zip) = 6832eb918ddf8f986fea206fa43a78b6. NOTE: This executable
was built in 2019, and thus the code-signing certificate has expired. First, you will need to accept that the disk is from an unknown publisher. Then
when you launch the app, you will see a message that "Windows protected your PC", with a button "Don't run". Instead, click on the "More info"
link, which will provide you with a button to "Run Anyway". Click that button. NOTE ALSO that some anti-virus programs may prevent the installer from running
with no indications that anything happened. If this occurs, you might need to choose "Run" from the Windows menu, type cmd into
the Open: text field to launch a command shell, type cd Desktop\BioTapestry7120,
and then type BioTapestryInstaller.exe. If an anti-virus program is activated (e.g. Symantec Endpoint Protection), you will then need
to allow the installer to run.
- Mac: There are now two different versions of BioTapestry for the Mac: one for Intel chips, and one for the newer Apple chips (i.e. M1 and M2 chips).
Be sure to download the version that matches your Mac!
Double-click the downloaded
dmg disk image file to mount it, and then open a Finder window for the mounted disk. From the Finder window,
drag the BioTapestry Editor icon inside onto your desktop (or, if you prefer, into your Applications folder). NOTE: If Mac Gatekeeper has been set to
restrict to only App Store, you will need to launch System Preferences, click on Security & Privacy, and on the General tab,
set Allow apps downloaded from: App Store and identified developers. If you take this step, you may wish to reset it back to App Store after
starting BioTapestry the first time.
-
Intel MD5 checksum:
MD5 (BioTapestryInstallImage713.dmg) = 442e1326923eef1014f72743f1c139fd
-
Apple (M1/M2) MD5 checksum:
MD5 (BioTapestryInstallImage713Mx.dmg) = 887d89adfa75f438946b68e4e19c82d2
- Linux: Compressed tar file: BioTapestry-7.1.2.0.tgz. Untar the file, which contains a
directory
BioTapestry. Inside that directory is the the shell script BioTapestry712.sh that launches the program. MD5 checksum: MD5 (BioTapestry-7.1.2.0.tgz) = 3335aa0af0c4118f73bbac87a5930358.
Getting Started
If you haven't used BioTapestry before, you can get up to speed on the basics by going through the online
Quick Start Tutorial.
Online GRN Repository
We have a hosted online repository of interactive BioTapestry gene regulatory network models at grns.biotapestry.org.
If you have a published model you would like us to host here at BioTapestry.org, contact us at biotapestry at systemsbiology dot org.
BioTapestry in Action
Here are some dynamic interactive BioTapestry models hosted at grns.biotapestry.org:
Interactive Web Models from the Davidson Lab
Other Interactive Web Models
Papers and Books Using BioTapestry
-
Xu, H., Chaudhri, V.K., Wu, Z., Biliouris, K., Dienger-Stambaugh, K., Rochman, Y., Singh, H.
Regulation of bifurcating B cell trajectories by mutual antagonism between transcription factors IRF4 and IRF8.
Nature Immunology, published online 05 October 2015. (Supplementary Materials)
-
Martik, M.L., McClay, D.R.
Deployment of a retinal determination gene network drives directed cell migration in the sea urchin embryo.
eLife 4:e08827, 2015.
-
Bouveret, R., Waardenberg, A.J., Schonrock, N., Ramialison, M., Doan, T., de Jong, D., Bondue, A., Kaur, G., Mohamed, S., Fonoudi, H., Chen, C.M., Wouters, M.A., Bhattacharya, S., Plachta, N., Dunwoodie, S.L., Chapman, G., Blanpain, C., Harvey, R.P.
NKX2-5 mutations causative for congenital heart disease retain functionality and are directed to hundreds of targets.
eLife 4:e06942, 2015.
-
Massie, C.E., Spiteri, I., Ross-Adams, H., Luxtonm H., Kay, J., Whitaker, H.C., Dunning, M.J., Lamb, A.D., Ramos-Montoya, A., Brewer, D.S., Cooper, C.S.,
Eeles, R., UK Prostate ICGC Group, Warren, A.Y., Tavaré, S., Neal, D.E., Lynch, A.G.
HES5 silencing is an early and recurrent change in prostate tumourigenesis.
Endocr Relat Cancer. 22(2):131-44, 2015.
-
Andrikou, C., Arnone, M.I.
Too many ways to make a muscle: Evolution of GRNs governing myogenesis.
Zoologischer Anzeiger - A Journal of Comparative Zoology 256: 2-13, 2015.
-
Simões-Costa, M., Bronner, M.E.
Establishing neural crest identity: a gene regulatory recipe.
Development 142(2): 242-257, 2015.
-
Andrikou, C., Pai, C.-Y., Su, Y.-H., Arnone, M.I.
Logics and properties of a genetic regulatory program that drives embryonic muscle development in an echinoderm.
eLife 4:e07343, 2015.
-
Peter, I.S., Davidson, E.H.,
Genomic Control Process: Development and Evolution, Academic Press, 2015.
-
Barsi, J.C., Li, E., Davidson, E.H.
Geometric control of ciliated band regulatory states in the sea urchin embryo.
Development 142(5): 953-61, 2015.
-
Imam, S., Noguera, D.R., Donohue. T.J.
An Integrated Approach to Reconstructing Genome-Scale Transcriptional Regulatory Networks.
PLoS Comput Biol. 11(2): e1004103, 2015.
-
Tonner, P.D., Pittman, A.M.C., Gulli, J.G., Sharma, K., Schmid, A.K.
A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea.
PLoS Genet. 11(1): e1004912, 2015.
-
Imam, S., Noguera, D.R., Donohue. T.J.
Global analysis of photosynthesis transcriptional regulatory networks.
PLoS Genet. 10(12):e1004837, 2014.
-
Cui, M., Siriwon, N., Li, E., Davidson, E.H., Peter, I.S.
Specific functions of the Wnt signaling system in gene regulatory networks throughout the early sea urchin embryo.
Proc Natl Acad Sci USA 111(47): E5029-38, 2014.
-
Li, E., Cui, M., Peter, I.S., Davidson, E.H.
Encoding regulatory state boundaries in the pregastrular oral ectoderm of the sea urchin embryo.
Proc Natl Acad Sci USA 111(10): E906-13, 2014.
-
Schwendinger-Schreck, J., Kang, Y., Holley, S.A.
Modeling the Zebrafish Segmentation Clock’s Gene Regulatory Network Constrained by Expression Data Suggests Evolutionary Transitions Between Oscillating and Nonoscillating Transcription
Genetics 197(2): 725–738, 2014.
-
Imam, S., Noguera, D.R., Donohue. T.J.
Global Analysis of Photosynthesis Transcriptional Regulatory Networks.
PLoS Genet. 10(12): e1004837, 2014.
-
Coleman, D.J., Garcia, G., Hyter, S., Jang, H.S., Chagani, S., Liang, X., Larue, L., Ganguli-Indra, G., Indra, A.K.
Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation.
PLoS Genet. 10(5): e1004321, 2014. (NOTE: See Figure S6 in supplemental materials.)
-
Singh, H., Khan, A.A., Dinner, A.R.
Gene regulatory networks in the immune system.
Trends in Immunology 35(5): 211-218, 2014.
-
Vander Lugt, B., Khan, A.A., Hackney, J.A., Agrawal, S., Lesch, J., Zhou, M., Lee, W.P., Park, S., Xu, M., DeVoss, J., Spooner, C.J., Chalouni, C., Delamarre, L., Mellman, I., Singh, H.
Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation.
Nature Immunology 15: 161–167, 2014.
-
Annunziata, R., Arnone, M.I.
A dynamic regulatory network explains ParaHox gene control of gut patterning in the sea urchin.
Development 141: 2462-2472, 2014.
-
Winck, F.V., Arvidsson, S., Riaño-Pachón, D.M., Hempel, S., Koseska, A., Nikoloski, Z., Urbina Gomez, D.A., Rupprecht, J., Mueller-Roeber, B.
Genome-Wide Identification of Regulatory Elements and Reconstruction of Gene Regulatory Networks of the Green Alga Chlamydomonas reinhardtii under Carbon Deprivation.
PLoS One 8(11): e79909, 2013.
-
Alcasabas, A.A., de Clare, M., Pir, P., Oliver, S.G.
Control analysis of the eukaryotic cell cycle using gene copy-number series in yeast tetraploids.
BMC Genomics 14: 744, 2013.
-
Streit, A., Tambalo, M., Chen, J., Grocott, T., Anwar, M., Sosinsky, A., Stern, C.D.
Experimental approaches for gene regulatory network construction: the chick as a model system.
Genesis 51(5): 296–310, 2013.
-
Paul, L., Wang, S.-H., Manivannan, S.N., Bonanno, L., Lewis, S., Austin, C.L., Simcox, A.
Dpp-induced Egfr signaling triggers postembryonic wing development in Drosophila.
Proc. Natl. Acad. Sci. USA 110(13): 5058–5063, 2013. (NOTE: See Figure S3 in supplemental materials.)
-
Hussey, S.G., Mizrachi, E., Creux, N.M., Myburg, A.A.
Navigating the transcriptional roadmap regulating plant secondary cell wall deposition.
Front Plant Sci. 4:325, 2013.
-
Ciau-Uitz, A., Pinheiro, P., Kirmizitas, A., Zuo, J., Patient, R.
VEGFA-dependent and -independent pathways synergise to drive Scl expression and initiate programming of the blood stem cell lineage in Xenopus.
Development 140: 2632-2642, 2013.
-
Gibb, N.L., Lavery, D.L., Hoppler, S.
sfrp1 promotes cardiomyocyte differentiation in Xenopus via negative-feedback regulation of Wnt signalling.
Development 140(7):1537-1549, 2013.
-
Aguilar-Hidalgo, D., Domínguez-Cejudo, M.A., Amore, G., Brockmann, A., Lemos, M.C., Córdoba, A., Casares, F.
A Hh-driven gene network controls specification, pattern and size of the Drosophila simple eyes
Development 140, 82-92, 2013.
-
Herrmann, F., Groß, A., Zhou, D., Kestler, H.A., Kühl, M.
A Boolean Model of the Cardiac Gene Regulatory Network Determining First and Second Heart Field Identity.
PLoS ONE 7(10): e46798, 2012.
-
Ogino, H., Ochi, H., Reza, H.M., Yasuda, K.
Transcription factors involved in lens development from the preplacodal ectoderm.
Dev Biol. 363(2): 333–347, 2012.
-
Zhang, Y., Yang, Y., Trujillo, C., Zhong, W., Leung, Y.F.
The expression of irx7 in the inner nuclear layer of zebrafish retina is essential for a proper retinal development and lamination.
PLoS One 7(4):e36145, 2012.
-
Metzakopian, E., Lin, W., Salmon-Divon, M., Dvinge, H., Andersson, E., Ericson, J., Perlmann, T., Whitsett, J.A., Bertone, P., Ang, S.L.
Genome-wide characterization of Foxa2 targets reveals upregulation of floor plate genes and repression of ventrolateral genes in midbrain dopaminergic progenitors.
Development 139(14):2625-34, 2012.
-
Jahangiri, L., Nelson, A.C., Wardle, F.C.
A cis-regulatory module upstream of deltaC regulated by Ntla and Tbx16 drives expression in the tailbud, presomitic mesoderm and somites.
Dev Biol. 371(1):110-20, 2012.
-
Luo, Y.J., Su, Y.H.
Opposing nodal and BMP signals regulate left-right asymmetry in the sea urchin larva.
PLoS Biol. 10(10):e1001402, 2012.
-
Kueh, H.Y., Rothenberg, E.V.
Regulatory gene network circuits underlying T cell development from multipotent progenitors.
Wiley Interdiscip Rev Syst Biol Med. 4(1):79-102, 2012.
-
Bessodes, N., Haillot, E., Duboc, V., Röttinger, E., Lahaye, F., Lepage, T.
Reciprocal signaling between the ectoderm and a mesendodermal left-right organizer directs left-right determination in the sea urchin embryo.
PLoS Genet. 8(12):e1003121, 2012.
-
Grocott T., Tambalo M., Streit A.
The peripheral sensory nervous system in the vertebrate head: A gene regulatory perspective.
Dev Biol. 370(1), 3-23, 2012.
-
Rabinowitz, A.H., Vokes, S.A.
Integration of the transcriptional networks regulating limb morphogenesis.
Dev Biol. 368(2), 165–180, 2012.
-
Woznica, A., Haeussler, M., Starobinska, E., Jemmett, J., Li, Y., Mount, D., Davidson, B.
Initial deployment of the cardiogenic gene regulatory network in the basal chordate, Ciona intestinalis.
Dev Biol. 368(1), 127–139, 2012.
-
Prasad, M.S., Sauka-Spengler, T., LaBonne, C.
Induction of the neural crest state: Control of stem cell attributes by gene regulatory, post-transcriptional and epigenetic interactions.
Dev Biol. 366(1), 10–21, 2012.
-
Peter, I.S., Davidson, E.H.
A gene regulatory network controlling the embryonic specification of endoderm.
Nature 474: 635-639, 2011.
-
Marsch-Martínez, N., Wu, W., de Folter, S.
The MADS symphonies of transcriptional regulation.
Front. Plant Sci. 2:26, 2011.
-
Owraghi, M., Broitman-Maduro, G., Luu, T., Roberson, H., Maduro, MF.
Roles of the Wnt effector
POP-1/TCF in the C. elegans endomesoderm specification gene network.
Dev Biol. 340(2), 209-221, 2010.
-
Saudemont, A., Haillot, E., Mekpoh, F., Bessodes, N., Quirin, M., Lapraz, F., Duboc, V., Röttinger, E., Range, R., Oisel, A., Besnardeau, L., Wincker, P., Lepage, T.
Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm.
PLoS Genet. 6(12):e1001259, 2010.
-
Eng, D., Campbell, A., Hilton, T., Leid, M., Gross, M.K., Kioussi, C.
Prediction of regulatory networks in mouse abdominal wall.
Gene 469(1-2):1-8, 2010.
-
Inoue, T., Sternberg, P.W.
C. elegans BED domain transcription
factor BED-3 controls lineage-specific cell proliferation during organogenesis.
Dev Biol. 338(2), 226-36, 2010.
-
Peter, I.S., Davidson, E.H.
The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage.
Dev Biol. 340(2): 188-199, 2010.
-
Danielli, A., Amore, G., Scarlato, V.
Built Shallow to Maintain Homeostasis and Persistent Infection:
Insight into the Transcriptional Regulatory Network of the Gastric Human Pathogen Helicobacter pylori.
PLoS Pathog. 6(6), e1000938, 2010. (NOTE: Bundled .btp file provided in supplemental materials.)
-
Baxter, L.L., Moreland, R.T., Nguyen, A.D., Wolfsberg, T.G., Pavan, W.J.
A curated online resource for SOX10 and pigment cell
molecular genetic pathways.
Database (Oxford) (2010), baq025, 2010.
-
Klucar, L., Stano, M., Hajduk, M.
phiSITE: database of gene regulation in bacteriophages.
Nucleic Acids Res. (38), D366-70, 2010.
-
Litvak, V., Ramsey, S.A., Rust, A.G., Zak, D.E., Kennedy, K.A., Lampano, A.E., Nykter, M., Shmulevich, I., Aderem, A.
Role of the transcription factor C/EBPδ in a regulatory circuit that discriminates between transient and persistent Toll-like receptor 4-induced signals.
Nat Immunol. 10(4): 437–443, 2009.
-
Pruszak, J., Isacson, O.
Molecular and Cellular Determinants for
Generating ES-Cell Derived Dopamine Neurons for Cell Therapy.
Adv Exp Med Biol. 651, 112-23, 2009.
-
Wahl, M.E., Hahn, J., Gora, K., Davidson, E.H., Oliveri, P.
The cis-regulatory system of the tbrain gene: Alternative use of multiple modules to
promote skeletogenic expression in the sea urchin embryo.
Dev Biol. 335(2), 428-41, 2009.
-
Kühn, C., Wierling, C., Kühn, A., Klipp, E., Panopoulou, G., Lehrach, H., Poustka, A.J.
Monte Carlo analysis of an ODE Model of the Sea Urchin Endomesoderm Network.
BMC Syst Biol. 3, 83, 2009.
-
Sansom, S.N., Griffiths, D.S., Faedo, A., Kleinjan, D.-J., Ruan, Y., Smith, J., van Heyningen, V., Rubenstein, J.L., Livesey, F.J.
The Level of the Transcription Factor Pax6 Is Essential for Controlling the
Balance between Neural Stem Cell Self-Renewal and Neurogenesis. PLoS Genet 5(6), e1000511, 2009.
-
Morohashi, K., Grotewold, E.
A systems approach reveals regulatory
circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 5(2), e1000396, 2009.
-
Su, Y.H., Li, E., Geiss, G.K., Longabaugh, W.J., Krämer, A., Davidson, E.H.
A perturbation model of the gene regulatory network for oral
and aboral ectoderm specification in the sea urchin embryo.
Dev. Biol. 329(2), 410-421, 2009.
-
Chan, T.-M., Longabaugh, W., Bolouri, H., Chen, H.-L., Tseng, W.-F., Chao, C.-H., Jang, T.-H., Lin, Y.-I., Hung, S.-C., Wang, H.-D., Yuh, C.-H.
Developmental gene regulatory networks in the zebrafish embryo.
Biochim. Biophys. Acta, 1789(4), 279-98, 2009.
-
Morley, R.H., Lachani, K., Keefe, D., Gilchrist, M.J., Flicek, P., Smith, J.C., Wardle, F.C.
A gene regulatory network directed by zebrafish No tail accounts
for its roles in mesoderm formation. Proc. Natl. Acad. Sci. USA 106, 3829-3834, 2009.
-
Bolouri, H.,
Computational Modeling of Gene Regulatory Networks: a Primer, Imperial College Press, 2008.
-
Tamplin, O.J., Kinzel, D., Cox, B.J., Bell, C.E., Rossant, J., Lickert, H.
Microarray analysis of Foxa2 mutant mouse embryos reveals
novel gene expression and inductive roles for the gastrula organizer and its derivatives. BMC Genomics 9:511, 2008.
-
Materna, S.C., Oliveri, P.
A protocol for unraveling gene regulatory networks. Nature Protocols 3(12), 1876-1887, 2008.
-
Smith, J., Davidson, E.H.
Gene regulatory network subcircuit controlling a dynamic
spatial pattern of signaling in the sea urchin embryo. Proc. Natl. Acad. Sci. USA 105, 20089-20094, 2008.
-
Nikitina, N., Sauka-Spengler, T., Bronner-Fraser, M.
Dissecting early regulatory relationships in the
lamprey neural crest gene network. Proc. Natl. Acad. Sci. USA 105, 20083-20088, 2008.
-
Ririe, T.O., Fernandes, J.S., Sternberg, P.W.
The Caenorhabditis elegans vulva: A post-embryonic
gene regulatory network controlling organogenesis. Proc. Natl. Acad. Sci. USA 105, 20095-20099, 2008.
-
Georgescu, C., Longabaugh, W.J.R., Scripture-Adams, D.D., David-Fung, E.-S., Yui, M.A., Zarnegar, M.A., Bolouri, H., Rothenberg, E.V.
A gene regulatory network armature for T lymphocyte specification. Proc. Natl. Acad. Sci. USA 105, 20100-20105, 2008.
-
Kioussi, C., Gross, M.K.
How to Build Transcriptional Network Models of Mammalian Pattern Formation. PLoS ONE 3(5), e2179, 2008.
-
Huang, T.S., Hsieh, J.Y., Wu, Y.H., Jen, C.H., Tsuang, Y.H., Chiou, S.H., Partanen, J., Anderson, H., Jaatinen, T., Yu, Y.H., Wang, H.W.
Functional network reconstruction reveals somatic stemness
genetic maps and dedifferentiation-like transcriptome reprogramming induced by GATA2.
Stem Cells 26(5), 1186-201, 2008.
-
Niessen, K., Karsan, A.
Notch Signaling in Cardiac Development. Circulation Research 102, 1169-1181, 2008.
-
Hoffman, B.G., Zavaglia, B., Witzsche, J., Ruiz de Algara, T., Beach, M., Hoodless, P.A., Jones, S.J.M., Marra, M.A., Helgason, C.D.
Identification of transcripts with enriched expression in the developing and adult pancreas. Genome Biology 9(6), R99, 2008.
-
Bonneau, R., Facciotti, M.T., Reiss, D.J., Schmid, A.K., Pan, M., Kaur, A., Thorsson, V., Shannon, P., Johnson, M.H., Bare, J.C., Longabaugh, W., Vuthoori, M., Whitehead, K.,
Madar, A., Suzuki, L., Mori, T., Chang, D.-E., DiRuggiero, J., Johnson, C.H., Hood, L., Baliga, N.S.
A predictive model for transcriptional
control of physiology in a free living cell. Cell 131(7), 1354-65, Dec 28 2007.
-
Vokes, S.A., Ji, H., McCuine, S., Tenzen, T., Giles, S., Zhong, S., Longabaugh, W.J.R., Davidson, E. H., Wong, W.H., McMahon, A.P.
Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development 134, 1977-1989, 2007.
-
Kühn, C., Kühn, A., Poustka, A.J., Klipp, E.
Modeling development: spikes of the sea urchin. Genome Inform. 18, 75-84, 2007.
-
Mori, A.D., Zhu, Y., Vahora, I., Nieman, B., Koshiba-Takeuchi, K., Davidson, L., Anne Pizard, A., Seidman, J.G., Seidman, C.E., Chen, X.J., Henkelman, R.M., Bruneau, B.G.
Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev. Biol. 297(2), 566-586, 2006.
-
Wellmer, F., Alves-Ferreira, M., Dubois, A., Riechmann, J.L., Meyerowitz, E.M.
Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genetics 2(7), e117, July 2006.
-
Livi, C.B., Davidson, E.H.
Regulation of spblimp1/krox1a, an alternatively transcribed isoform expressed in midgut and hindgut of the sea urchin gastrula.
Gene Expr. Patterns 7(1-2), 1-7, 2007.
-
Livi, C.B., Davidson, E.H.
Expression and function of blimp1/krox, an alternatively transcribed regulatory gene of the sea urchin endomesoderm network. Dev. Biol. 293(2), 513-25, 2006.
-
Davidson, E.H., Erwin, D.H.
Gene regulatory networks and the evolution of animal body plans. Science 311 (5762), 796-800, 2006.
-
Davidson, E.H.,
The Regulatory Genome: Gene Regulatory Networks in Development and Evolution, Elsevier, 2006.
-
Koshiba-Takeuchi, K., Takeuchi, J.K., Arruda, E.P., Kathiriya, I.S., Mo, R., Hui, C-c., Srivastava, D., Bruneau, B.G.
Cooperative and antagonistic interactions between Sall4 and Tbx5 pattern the mouse limb and heart. Nature Genetics 38, 175-183, 2005.
-
Levine, M., Davidson, E.H.
Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 102, 4936-4942, 2005.
-
Stathopoulos A., Levine M.
Genomic regulatory networks and animal development. Dev Cell. 9(4), 449-62, Oct 2005.
Please let us know if you are using BioTapestry for your research!
Description
BioTapestry is designed around the
concept of a developmental network model, and is intended to deal
with large scale models with consistency and clarity.
It is capable of representing systems that exhibit
increasing complexity over time, such as the genetic regulatory
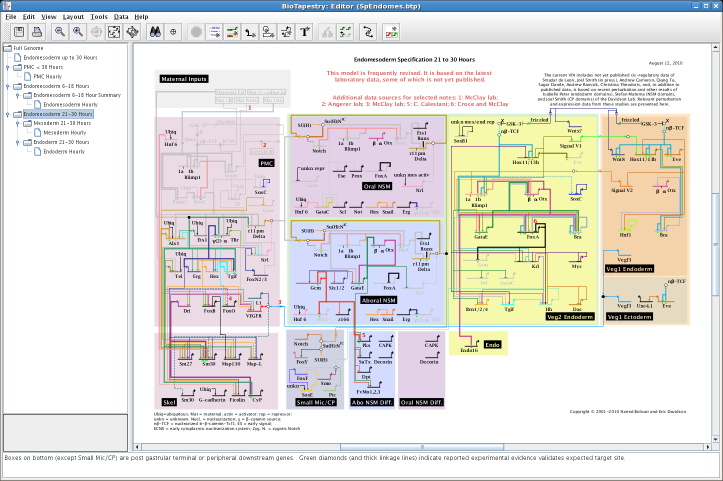
network controlling endomesoderm development in sea urchin embryos:

Another system that demonstrates BioTapestry's capabilities for representing systems that exhibit
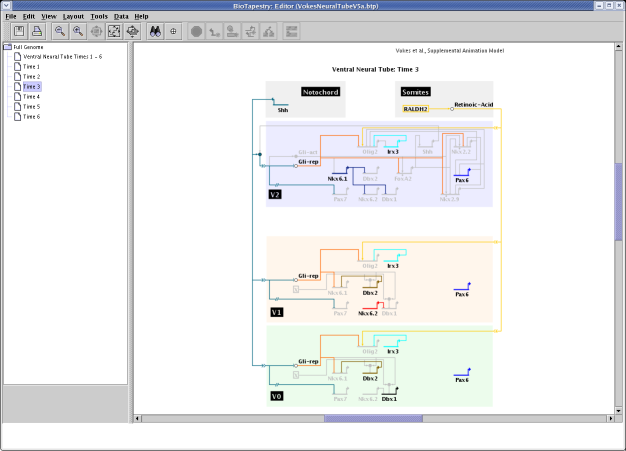
increasing complexity over time is the network for mouse ventral neural tube specification (Vokes et.al., 2007):
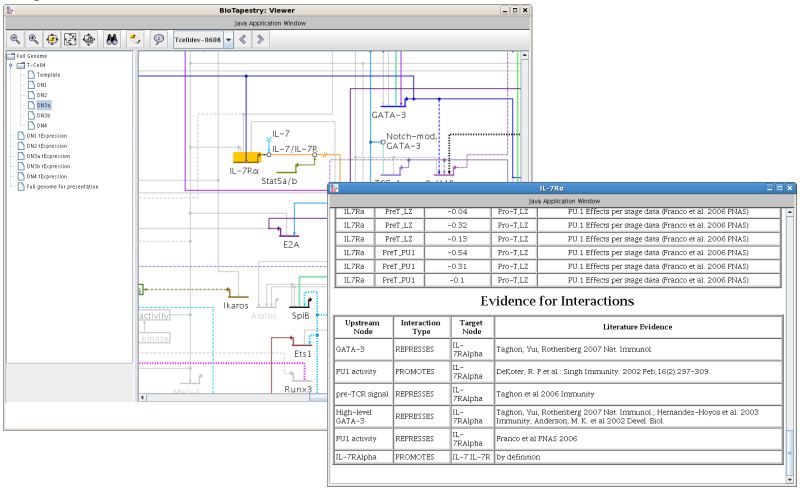
Users can annotate network elements with the supporting experimental evidence, as is shown here for the T-cell gene regulatory network
(Georgescu et. al., 2008):
BioTapestry's auto layout templates and resizable workspace make it a good tool for working with much larger networks, such as this network
for Halobacterium salinarum (Bonneau et.al., 2007):
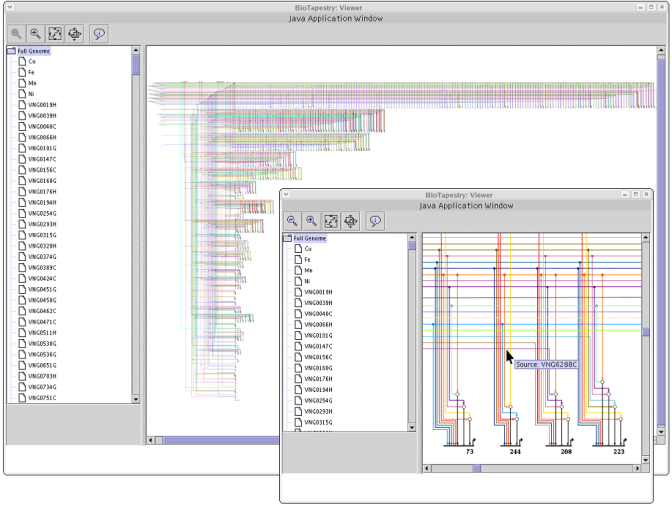
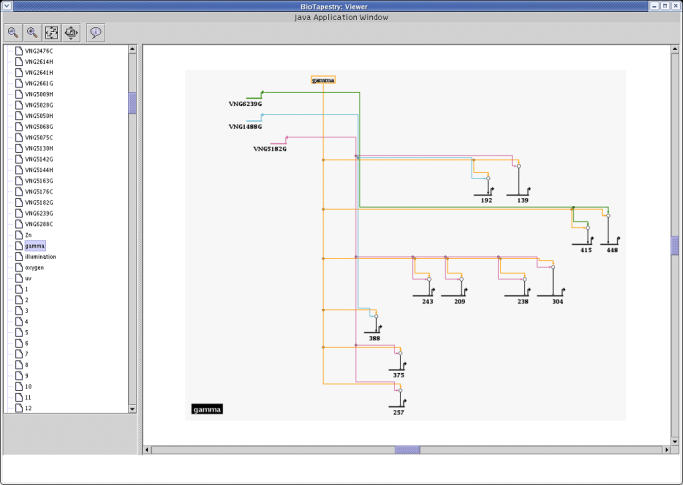
BioTapestry's framework for creating sets of submodels is well-suited for organizing such large networks in an understandable way, as this detail from the above
Halobacterium salinarum network demonstrates:
Significant features include:
-
Supporting data resulting from the perturbation of expression of
specific genes, measured in any way (QPCR, genetics, etc.), can
be easily accessed for each gene or link. Temporal and spatial
expression results are also accessible.
-
BioTapestry can portray hourly, localized views of the network
during development based on data tables describing the network's
local and temporal states.
-
BioTapestry can automatically layout the set of network models from
lists of interactions provided either through interactive dialogs
or comma-separated value (CSV) files exported by spreadsheet programs.
Running BioTapestry Online Using Java Web Start
Running BioTapestry as a Java Web Start application is no longer supported; please use the downloadable versions of the program provided above, which
all include the required Java components needed to run BioTapestry. Our apologies for any inconvenience.
Documentation
BioTapestry Frequently Asked Questions
If you have a question about how something works in BioTapestry, consult the online
BioTapestry Frequently Asked Questions list (though be advised it
has not been updated in quite a while). If you have a question that isn't covered there,
then please let us know so we can add it to the list.
Online Tutorials
A series of short tutorials that focus on different aspects of the software are available online.
For example, for newcomers, the Quick Start Tutorial covers the
minimum material you need to get started working with BioTapestry. There are other tutorials that build
upon that introductory material and cover more advanced topics.
Current Online Tutorial List
Quick Start Tutorial: This is the best way to learn the basics for
using BioTapestry. It covers the minimum material you need to draw a multi-level hierarchy of network models.
If you are a newcomer, this will be the best way to begin.
| Quick Start Tutorial Resource |
File (right-click & use "Save Link As...") |
| Completed BioTapestry Model | QuickStartModel.btp |
(The original Revision A of the Quick Start Tutorial is still available here
if you want to learn about how to build networks using the original "top-down" method.)
Dynamic Submodels Tutorial: This tutorial covers how to create dynamic submodels,
where data tables are used to automatically determine the active network elements, and the user can manipulate a time slider to see a
dynamic presentation of the network behavior. This is the approach used for the S. purpuratus endomesoderm model.
Tutorial on Building Networks from Interaction Tables: This tutorial covers
how to build and layout networks automatically from lists of interactions, which are specified using interactive dialogs.
It also covers how to use several tools that can help with common network layout tasks; these are useful even if you are
drawing all your networks manually.
Building Networks from Comma-Separated Value Files: This tutorial covers
how to build and layout networks automatically using lists of interaction commands that are created in a spreadsheet
program such as Microsoft Excel, and then imported into BioTapestry as a comma-separated value (CSV) file.
Tutorial on Using and Creating Paths: This tutorial covers the
feature of BioTapestry that allows you to define named paths through the model hierarchy. You may have
situations where you have a complex model hierarchy, and you want to be able to easily progress through a series of closely
related submodels that could be widely dispersed across that hierarchy. Paths allow you to do this.
What's New
Version 7.1.3 was released on 9/25/23. This version is only for Mac computers. The only significant difference is that the code has been notarized by Apple
to ease installation, and that a version of Java that runs on Apple M1 and M2 chips has been incorporated into a different executable. The feature set is
identical to the 7.1.2 Windows and Linux versions; those executables remain the current versions to install on those platforms, despite the fact the the code
signing certificate has now expired for those platforms. The certificate, verifying that the software originates from the Intitute for Systems Biology, was current
at the time the software was released (9/28/19).
Version 7.1.2 was released on 9/28/19, and the only notable differences from the prior release are that: 1) all platforms bundle the
required Java components; 2) the Windows version is now installed using an install program; and 3) all releases are signed with a current
(as of 9/28/19) signing certificate verifying that the software originates from the Institute for Systems Biology.
Version 7.1.1 was released on 9/24/17, and is a minor bugfix release. The one significant change is that
the version for the Mac is now signed with an Apple Developer Certificate, and also has the Java
Runtime embedded in the application; you do not have to download and install Java separately for the Mac.
Version 7.1.0 was released on 9/29/16, and is an interim bugfix release. Some useful new features have been added
as well, which are described in detail in the Version 7.1 Release Notes.
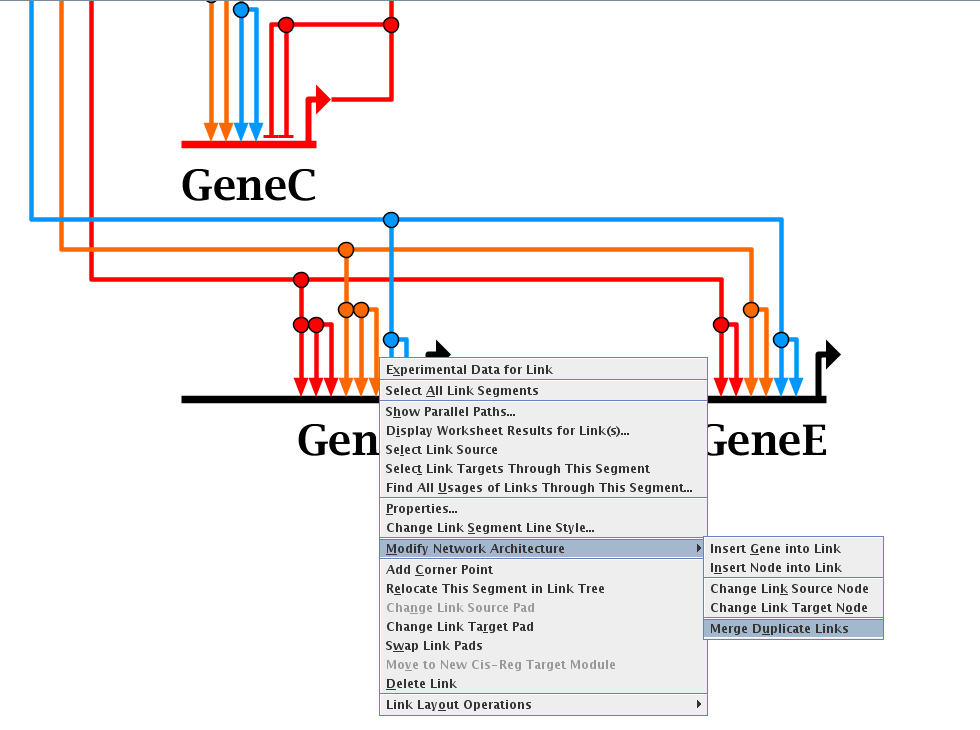
One set of new features was designed to help clean up some of the unneeded or duplicated elements that can commonly accumulate in
the top-level model after the user builds their models at lower levels in the hierarchy. For example, there can be duplicate links or nodes, or
unused elements, and new tools have been added to track down and eliminate these unnecessary elements:
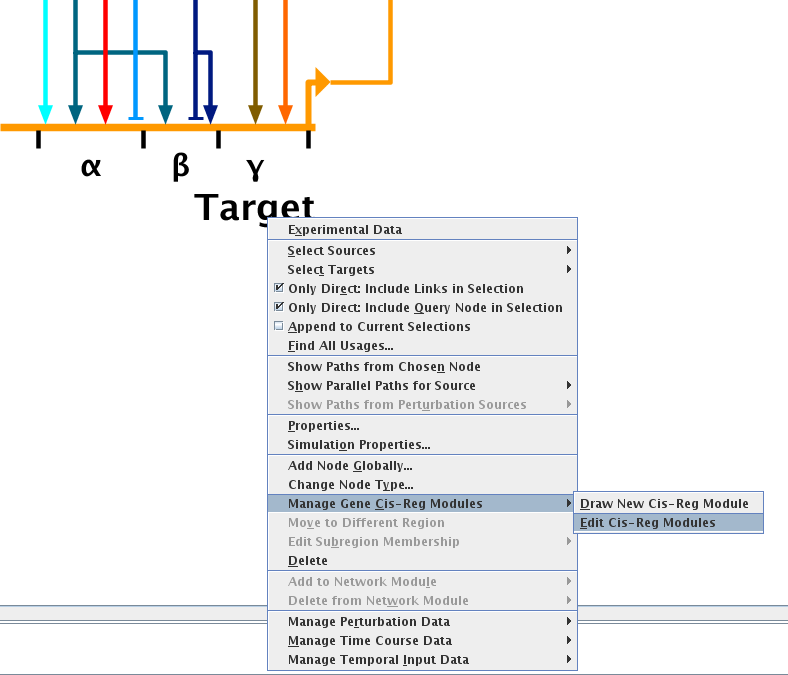
Version 7.1 has also made it much easier to create, edit, and maintain cis-regulatory element definitions for genes. The modules can now be drawn instead of
specified in an obscure table, and an editing dialog replaces the need to change table entries. One of the most significant improvements is that links are now matched with their target modules, and the system
will manage links correctly as cis-reg modules are reordered, resized, and shifted.
More in-depth discussion of these new features can be found in the Version 7.1 Release Notes.
Older release notes are also available for Version 7,
Version 6,
Version 5,
Version 4,
Version 3,
Version 2 and for even
older versions.
Publications
Please cite BioTapestry using [Longabaugh et.al. 2005],
[Longabaugh et.al. 2009],
and/or [Paquette et.al. 2016] if you use BioTapestry in your research.

-
Paquette, S.M., Leinonen, K., Longabaugh, W.J.R.
BioTapestry now provides a web application and improved drawing and layout tools [version 1; referees: 3 approved].
F1000Research 5:39, 2016.
-
Longabaugh, W.J.R.
BioTapestry: A Tool to Visualize the Dynamic Properties of Gene Regulatory Networks.
Methods Mol Biol. 786, 359-94, 2012.
-
Longabaugh, W.J.R., Davidson, E.H., Bolouri, H.
Visualization, documentation, analysis, and communication of large-scale gene regulatory networks.
Biochim. Biophys. Acta, 1789(4), 363-74, 2009.
-
Longabaugh, W., Bolouri, H.
Understanding the Dynamic Behavior of Genetic Regulatory Networks by Functional Decomposition. Current Genomics, Volume 7, Number 6, 333-341, 2006.
-
Longabaugh, W.J.R., Davidson, E.H., Bolouri, H.
Computational representation of developmental genetic regulatory networks.
Dev. Biol. 283, 1-16, 2005.
Problems Running It?
If you run into any issues with getting BioTapestry to run on your computer that you cannot
resolve, or have a program crash, please send a report to biotapestry at systemsbiology dot org or post a message
on the BioTapestry-users Google group
so we can address your problem. Browsing the Google group is a good way of finding out about any
recently identified compatibilty issues and what workarounds have been developed to addess them.
If you have previous BioTapestry files that are not loading into the current version correctly,
or run into any other show-stopping bugs with the new version, the previous versions (e.g. 7.0.0, 6.0.0) can
still be dowloaded from the Archived Executables page.
Supported Platforms
There are BioTapestry downloadable executables for Windows, Mac, and Linux. All three versions require 64-bit versions
of your operating system. Thus, 32-bit Windows systems are no longer supported. If you have a 32-bit Windows system, you
will need to download Version 7.1.1 (or older) of BioTapestry from the Archived Executables page
and also install a 32-bit version of Java; see here. Note that Oracle
has recently changed Java licensing, which could be an issue for you, given your particular circumstances.
BioTapestry for Gaggle
Beginning with Version 7.1.0, we are no longer providing updates to the Gaggle-enabled BioTapestry Editor. Instructions and the
final release Version 7.0.0 Web Start link are provided here.
Developer's Notes
BioTapestry supports user-written plugins for creating a customized
experimental data display. There are example plugins in the
source code tree (see below) that can serve as a starting point.
If you need any guidance on building plugins, feel free to
contact us.
BioTapestry can be called in batch mode from within another Java program to
generate images from a comma-separated-value specification coming from a
provided InputStream. A small example is provided in the source code tree
(see below) in the org.systemsbiology.biotapestry.app.ImageGeneratorAppTestWrapper
class.
The BioTapestry Viewer can be embedded as a visualization component inside another Java program.
An example of how to do this is shown in the class
org.systemsbiology.biotapestry.embedded.EmbeddedViewerPanelTestWrapper in the source code.
Source Code
Starting with Version 7, BioTapestry source code is now hosted on GitHub. There are
separate repositories; one for the Java codebase and another for the
new browser-based Viewer Web Application code. A snapshot of the code
for version 7.1.2 is available on the GitHub 7.1.2 release page.
BioTapestry Viewer
BioTapestry has always supported the ability to share interactive, read-only GRN models over the web.
Prior to Version 7, the BioTapestry Viewer used Java Web Start to run on the user's computer. We now support a new way of hosting a
Viewer embedded in the web browser that does not require the user to have Java installed on their computer. This change requires that the
web server hosting the Viewer supports Java Servlets (e.g. Apache Tomcat Version 6). We provide a page with detailed instructions
on how to get a Viewer installation up and running. You can also contact us using the resources listed in the Feedback section
if you are having problems.
If your deployment is on a publically available web site, please let us know
about it. We'd like to link to your viewer page off the BioTapestry home page.
Even if you prefer not to be linked to, we appreciate being able to keep track of viewer
deployments. Thanks!
Send Feedback
We are eager to receive any feedback you wish to send us, such
as questions, bug reports, feature requests, and usability issues.
The best place to do this is in the
BioTapestry-users Google group.
However, you can also email us privately at:
biotapestry at systemsbiology dot org
BioTapestry-users Google Group
This group is intended to give BioTapestry users a place to discuss
the program, get help with questions, ask for program enhancements,
provide feedback, and share tips with each other on using the
software. The group home page is:
https://groups.google.com/group/biotapestry-users
Credits
BioTapestry began as a joint project of the Institute for Systems Biology (ISB) and the Davidson Lab at Caltech. The orginal contributors were:
- William Longabaugh
- Eric Davidson
- Hamid Bolouri
Since 2012, all program development is solely based at ISB, under the direction of William Longabaugh. From Autumn 2012 to Spring 2016,
the BioTapestry development team at ISB also included:
- Suzanne Paquette
- Kalle Leinonen
Also:
- Stephen Ramsey, formerly at the Institute for Systems Biology, helped to write the
SBML output and simulation support
- Dan Tenenbaum, formerly at the Institute for Systems Biology, helped to write the
Gaggle 2007-4 support
Windows executable created using the Launch4j Executable Wrapper by Grzegorz Kowal
Code is available on Sourceforge.
The Mac executable was formerly bundled with the appbundler code provided by Oracle and Infinite Kind. Starting with Version 7.1.3,
it is packaged using the jpackage utility bundled with the Java JDK.
BioTapestry development was previously supported by the National Institute of General Medical Sciences under Award
Number R01GM061005, and the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the
National Institutes of Health under Award Number R01HD073113. The content of this web site is
solely the responsibility of the authors and does not necessarily represent the official views of the National
Institutes of Health.
License and Disclaimer
BioTapestry is Copyright (C) 2003-2023, Institute for Systems Biology.
This software is licensed under the GNU Lesser General Public License (LGPL), which is a standard "free software"
and "open source" license.
Some of the toolbar icon images in this package are covered by a free distribution license from Sun Microsystems.
The downloadable Windows executable is wrapped in code that is made available by Grzegorz Kowal under
an MIT License.
The Windows installer uses the Nullsoft Scriptable Install System (NSIS), distributed under this license.
The downloadable Mac executable is bundled with the appbundler code that is copyright Oracle, and is licensed under the
GNU General Public License version 2 with "Classpath" exception. Additional code in that
bundle is copyright Infinite Kind, and is also licensed under the same GNU General Public License version 2 with "Classpath" exception.
Executables for all platforms include a Java Runtime Environment built from the OpenJDK by Oracle, which is covered by the
GNU General Public License Version 2 with "Classpath" exception. Some parts of the OpenJDK are
also subject to terms described in Additional License Info and
Assembly Exception. Specific license terms for those parts are included in the legal folder
of each bundled Java runtime.
Despite our best efforts, there are bugs in BioTapestry. Please be advised that, as stated in the
license agreement, the Institute for Systems Biology and the authors disclaim any
liability stemming from the use of this software.
|












